Molecular-level Intelligence Evidenced by Discovery of Extensive Adaptive RNA Editing in Octopus Transcriptome

Alienesque cephalopods just got even more interesting: scientists have found that octopuses can recode their transcriptome to optimize neuronal function under differing environmental conditions.
By: William Brown, scientist at the Resonance Science Foundation
This article has an accompanying video review:
The Physics of Intelligence
A significant achievement of science over the past few hundred years has been in removing supernatural and magical explanations of natural phenomena and inculcating within the modern mind the ability to conceptualize natural phenomena as fully mechanical processes. This has enabled us to understand myriad natural phenomena with mathematical precision. From this progress, it is now the predominant and conventional perspective to view all processes as fundamentally mechanistic. This has an interesting ramification on the subject of natural intelligence, as in the canon of conventional scientific theory, there is no innate intelligence within the universe: you have systems that replicate intelligence, via mechanics, but those natural systems are essentially no different from man-made machines that may have functions of the machinery that seem intelligent, but are fundamentally lifeless and blind.
In cases where there are observable uncanny intelligence, like the biological system, the conventional scientific mind will evoke computation; the brain is computational (a machine) that produces behaviors, some which might seem volitional, but they are fundamentally deterministic and predictable from the programming of the machine (in this case the neuronal hardwiring of the brain-as-a-computer model). As well there are genetic algorithms that computationally determine evolutionary traits, and seemingly intelligent behaviors of animals are mere instincts programmed by the genetic circuitry, i.e., animals do not actually possess intelligence; such behavior seeming to exhibit innate intelligence is only superficial and at a core level is simulated from underlying predictable programmed genetic algorithms.
So, mechanics and computation generate an intelligence in the universe that is in effect, superficial. One of the primary reasons for the strong resistance of science to accepting innate intelligence, perhaps, is that it opens the door to perceiving natural sentience, which leads to questions of teleology, and this in turn invites philosophical or even spiritual perspectives that hard science wants to avoid or ignore. But these are not the only two alternatives, there is a scientific way to understand innate intelligence that does not invite magical thinking but views the intrinsic and integral connectivity of all systems as a self-organizational dynamic that engenders within it a natural sentience and volition.
At the fundamental-most scale the universe is a singular quantum field, and there is a continual flux of energy and information through this field. This flux of information, as it is coordinated through the multiply connected geometry of the entanglement network of space (spacememory), animates the universe with qualities of an innate intelligence present in the self-organizational dynamics, in fact a hyperintelligence and natural sentience, that is not the result of computation or clockwork, but due to the interrelationship (often nonlocal) between all subsystems of the universe, which synergistically self-organize in a hyper-intelligent fully cooperative and inter-connected system.
In exploration of this key concept, we will explore here the understanding that intelligence in the biological system is not restricted to a complex central nervous system, like the brain. Processing of environmental information and information of internal states— generating intelligent directionality of cellular operations—resides at a much more primordial level than neuronal networks, at the heart of the cell in the genetic, morphogenic, and metabolic molecular networks of the cell, which necessarily operate at the quantum level and hence involve nonlocal dynamics of the wavefunctions.
Since the demonstrable intelligence we observe in neuronal activity evolutionarily emerges from the intelligence of the molecular networks of the cell, which possesses the same quality of information processing normally only attributed to entire neuronal networks, as a result of being a holo-fractal system with scale-free cognitive function, we should not be surprised to find intelligence operational within the genetic machinery of the living system, as it is the morphogenic networks— the trans-temporal information exchange in the spacememory network— that are the ultimate root of intelligence in the organism, and as we have postulated in previous work, this hyperintelligence is present even in the biochemical networks and genetic circuits of the living system.
In a direct observation of this molecular-level intelligence a recent study has discovered extensive adaptive RNA editing in the octopus’ transcriptome in response to environmental conditions. The research shows that cephalopods can adapt to changes in temperature by tweaking their genetic code to alter the proteins that are made in their nerve cells.
Temperature-dependent RNA editing in cephalopods extensively recodes the neural proteome
We have all heard of the genome, which is relatively (though not entirely [1]) set at inception with the code and genetic cassettes that an organism will have for its lifespan. There is a complimentary body of genetic code that—while not in the limelight as often—is more dynamic than the genome where much of the action of turning a genotype into a phenotype occurs, and this is referred to as the transcriptome. The genes encoded by the nucleotide base pair sequences of DNA are transcribed into intermediary RNA messenger transcripts (mRNA) forming the transcriptome where many gene-editing events take place before processed mRNA transcripts are exported from the nucleus to the cytoplasm where polypeptide synthesis machinery of the ribosome can access the code to translate it into the specified protein.
RNA editing is one such processing event, a class of epigenetic modification, and occurs when the nucleotide code is changed; for instance, editing an adenosine base to an inosine base, which is read differently by the ribosome-tRNA translation machinery. Such RNA editing is a widespread epigenetic process that can alter the amino acid sequence of proteins and is termed “recoding”. It has been discovered that in cephalopods—more so than any other species studied so far— most transcripts are recoded, and recoding is hypothesized to be an adaptive strategy to generate phenotypic plasticity. Adenosine to inosine (A-to-I) editing occurs widely across animals and serves many functions, including the regulation and diversification of mRNAs [2].
In most animals, recoding through RNA editing is relatively rare; for example, recoding constitutes less than 1% of all reported editing sites in humans [3]. In contrast, RNA recoding occurs extensively in soft-bodied cephalopods [4]: 60% of all mRNAs are recoded in squid, octopus, and cuttlefish [5]. RNA sequencing has revealed tens of thousands of recoding sites across the transcriptomes of these animals, and most recoded transcripts harbor multiple recoding sites. RNA recoding in cephalopods therefore has the potential to dramatically diversify the proteome. Although not nearly as extensive as cephalopods, the only other organism with a comparable level of recoding is humans, specifically in the neuronal information processing tissue. So, it is interesting that despite the evolutionary distance between humans and cephalopods, which are veritable alien organisms compared to our biology, there is convergent evolution of RNA gene editing networks that are directly correlated with higher-order cognitive capabilities [6]. The demonstrable intelligence of these two species is rooted in a molecular-level intelligence of environmental sensing with rapidly adaptive RNA editing and proteome plasticity.

Figure 1. Cephalopods activate A-to-I RNA editing to epigenetically modify mRNA transcripts and generate novel protein vairants. RNA recoding in squid specifies unique kinesin variants with distinct activities in different tissues and in response to changes in seawater temperature, and cephalopod recoding sites provide a guide to identifying functional substitutions in non-cephalopod motor proteins. RNA editing in squid specifies unique kinesin protein variants in different tissues. Unique kinesin variants are made acutely in response to seawater temperature. Cold-specific kinesin variants have enhanced single molecule motility in the cold. Cephalopod editomes (body of editing mRNA transcripts) can reveal functional substitutions in non-cephalopod proteins. Image and image description from: [4] K. J. Rangan and S. L. Reck-Peterson, “RNA recoding in cephalopods tailors microtubule motor protein function,” Cell, vol. 186, no. 12, pp. 2531-2543.e11, Jun. 2023.
In a new study [7], Joshua Rosenthal at the Marine Biological Laboratory in Woods Hole, Massachusetts, and his colleagues tested how California two-spot octopuses (Octopus bimaculoides) responded to changes in water temperature in tanks. They gradually shifted the temperature to around 13°C (55°F) for one group and 22°C (72°F) for another group. The octopuses in the colder tank made more than 13,000 edits to their RNA that led to changes in the resulting proteins that are vital for neural processes.
For two highly temperature-sensitive examples, it was discovered that recoding tunes protein function, a molecular-level intelligent response. One such product variant that was identified was a neuronal protein called synaptotagmin: a key component of Ca2+-dependent neurotransmitter release. In the study, crystal structures and supporting experiments revealed that editing alters Ca2+ binding, optimizing neurotransmitter release for given environmental temperatures. The second protein, also integrally involved in neurophysiology, is kinesin-1—a motor protein driving axonal transport along the core microtubule filamentary bundle—editing regulates transport velocity down microtubules. Seasonal sampling of wild-caught specimens indicates that temperature dependent editing occurs in the field as well. These data show that A-to-I editing tunes neurophysiological function in response to temperature in octopus and most likely other cephalopods.
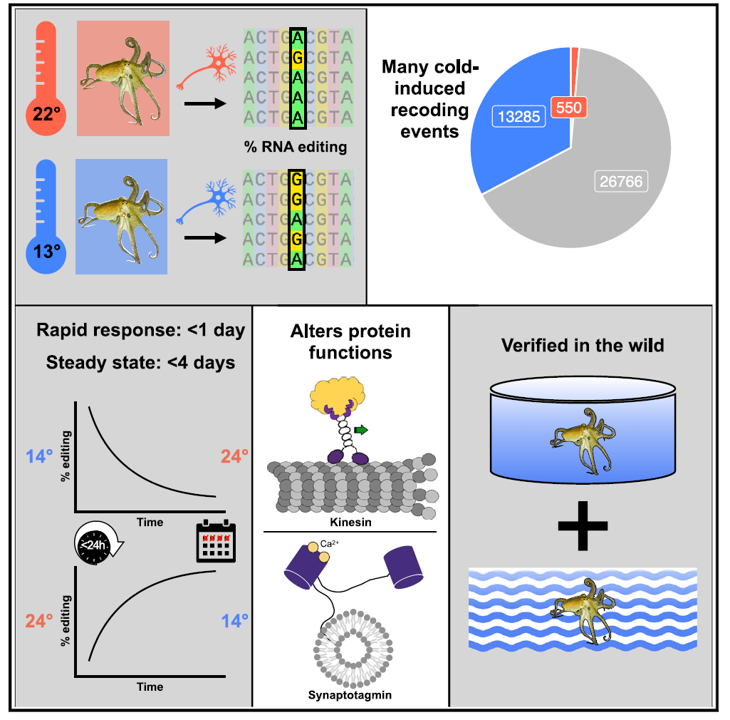
Figure 2. Octopuses utilize RNA editing to rapidly respond to environmental temperature changes by altering protein function. Octopus bimaculoides increase A-to-I RNA editing at >13,000 sites when in cold temperatures. Editing shifts occur within hours and are observed in wild populations. As a functional example, one cold-induced site alters kinesin motility, another cold-induced site alters Ca2+-binding affinity in vesicle fusion protein synaptotagmin. Image and image description from: [7] M. A. Birk et al., “Temperature-dependent RNA editing in octopus extensively recodes the neural proteome,” Cell, vol. 186, no. 12, pp. 2544-2555.e13, Jun. 2023.
Unified Science- In Perspective
In our Unified Science articles we often focus on experiments and theories advancing unified physics, which is a mathematical and conceptual unification of all forces, constants, properties, dynamics and interactions of matter and energy from the quantum to the cosmological scale (see Nassim Haramein’s and Olivier Alirol’s scale invariant unification research). However, we are interested in much more than just the field of physics; one of the reasons that we refer to unified science is because we are actively extending the concept of unified physics to include a unified description of all organized matter in the universe, which extends beyond just quantum physics and relativity and into chemistry, biology, psychology, complex systems, and intelligence in the universe.
So, for example, we can look at the dynamics of quantum gravity operational in the biological system [8], where the underlying spacetime geometry of quantum entangled wavefunctions of macromolecules direct the biochemical networks of the cell and the coupled information exchange of the biological system with the universal information network of spacememory. Where nonlocal signals are quasi-instantaneously exchanged regardless of distance in space or time via micro-wormholes manifesting as quantum tunneling dynamics. Our work to unify science may be a bit ahead of its time, as more conventional scientists most probably view quantum gravity and biology as two separate and distinct worlds, having nothing to do with each other. Of course, this is the fallacy of a myopic and non-syncretic perspective, and time and observational science and experiment will, in time, come to show the indelible and innate link between these two domains, and indeed the innate interconnection of all domains within the universe.
In our work on the Unified Spacememory Network one of our primary postulates is that evolution involves non-random mechanisms that have directionality as a function of the constitutive information flux through the connectivity architecture of spacememory underlying self-organizational dynamics and system intelligence. An example of this is the information feedback-feedforward and memory processes occurring within the molecular domain of the living system, and within the unified physics domain as the molecular dynamics and organization are linked with the information processes of the spacememory network, which gives the molecular domain of the biological system a hyperintelligence.
So, an organism can direct evolutionary adaptations, not because at the organism level it is cognitively directing these changes, but because life is a nested system of cognitive domains, there is scale-free cognition. We extend this across all scales, so even biochemical networks have cognitive-type information processing, or innate intelligence. This is how you get biogenesis; how at one point, probably early on in the history of the universe, certain gestalts of abiotic matter transitioned to living matter, primordial chemical networks were not blind and / or random, directional adaptation was able to proceed in an intelligent fashion to convert some biochemistry to a medium of memory with information coding and syntax (what we know today as DNA), and direct cooperative networks that formed compound intelligences, like the first cell, and then multicellular organisms and onwards.
This latest study and revelations of the widespread intelligent and directionally adaptive RNA editing occurring in the cephalopod transcriptome is an empirical demonstration of the molecular-level intelligence of the biological organism and the nested-holofractal organization of scale-free cognition that quintessentially characterizes biology. In science novel postulates are the beginnings of new theories, and we are glad to see our arbeitshypothese as delineated in the Unified Spcaememory Network and elsewhere seeing empirical validation, because it is the crucible of experimentation and testing that advances a hypothesis to a theory and finally to a working model. In the larger purview, experiments and results such as what was discovered in this latest study of cephalopod molecular-level intelligence will show that the living organism is not a simple molecular automaton—which is the predominant perspective / assumption in today’s conventional scientific paradigm. But instead, that the scale-free cognition and holo-fractal organizational dynamics of the biological system endow it with a volitional character beyond programming and pre-deterministic mechanics: see my preprint article [9] to take a deeper dive into this idea- Provisional Definition of the Living State: Delineation of an Empirical Criterion that Defines a System as Alive.
References
[1] S. Bachiller, Y. del-Pozo-Martín, and Á. M. Carrión, “L1 retrotransposition alters the hippocampal genomic landscape enabling memory formation,” Brain, Behavior, and Immunity, vol. 64, pp. 65–70, Aug. 2017, doi: 10.1016/j.bbi.2016.12.018
[2] Nishikura, K. (2016). A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96. https://doi.org/10.1038/nrm.2015.4
[3] Gabay, O., Shoshan, Y., Kopel, E., Ben-Zvi, U., Mann, T.D., Bressler, N., Cohen-Fultheim, R., Schaffer, A.A., Roth, S.H., Tzur, Z., et al. (2022). Landscape of adenosine-to-inosine RNA recoding across human tissues. Nat. Commun. 13, 1184. https://doi.org/10.1038/s41467-022-28841-4
[4] K. J. Rangan and S. L. Reck-Peterson, “RNA recoding in cephalopods tailors microtubule motor protein function,” Cell, vol. 186, no. 12, pp. 2531-2543.e11, Jun. 2023, doi: 10.1016/j.cell.2023.04.032
[5] Rosenthal, J.J.C., and Eisenberg, E. (2023). Extensive Recoding of the Neural Proteome in Cephalopods by RNA Editing. Annu. Rev. Anim. Biosci.11, 57–75. https://doi.org/10.1146/annurev-animal-060322-114534
[6] Brown, W.D. Convergent Function of Retrotransposons in Octopus Brain Drive Sophisticated Cognitive Capabilities. The Resonance Science Foundation. July 11, 2022. https://www.resonancescience.org/blog/convergent-function-of-retrotransposons-in-octopus-brain-drive-sophisticated-cognitive-capabilities
[7] M. A. Birk et al., “Temperature-dependent RNA editing in octopus extensively recodes the neural proteome,” Cell, vol. 186, no. 12, pp. 2544-2555.e13, Jun. 2023, doi: 10.1016/j.cell.2023.05.004
[8] N. Haramein, W. D. Brown, and A. Val Baker, “The Unified Spacememory Network: from Cosmogenesis to Consciousness,” Neuroquantology, vol. 14, no. 4, Jun. 2016, doi: 10.14704/nq.2016.14.4.961
[9] W. Brown, “Provisional Definition of the Living State: Delineation of an Empirical Criterion that Defines a System as Alive,” preprint, Jul. 2023. doi: 10.32388/V5EDGF



